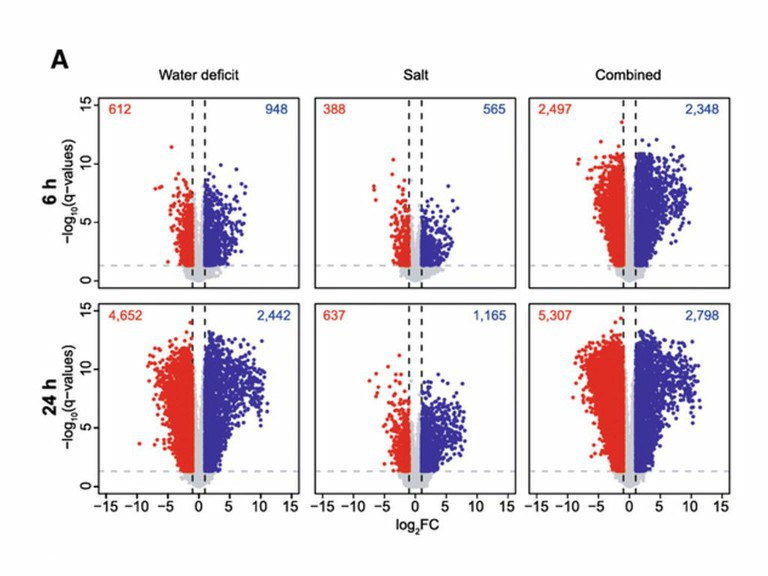
Water deficit and soil salinity substantially influence plant growth and productivity. When occurring individually, plants often exhibit reduced growth resulting in yield losses. The simultaneous occurrence of these stresses enhances their negative effects. Unraveling the molecular mechanisms of combined abiotic stress responses is essential to secure crop productivity under unfavorable environmental conditions.This study examines the effects of water deficit, salinity and a combination of both on growth and transcriptome plasticity of barley seminal roots by RNA-Seq. Exposure to water deficit and combined stress for more than 4 days significantly reduced total seminal root length. Transcriptome sequencing demonstrated that 60 to 80% of stresstype-specific gene expression responses observed 6 h after treatment were also present after 24 h of stress application. However, after 24 h of stress application, hundreds of additional genes were stress-regulated compared to the short 6 htreatment. Combined salt and water deficit stress application results in a unique transcriptomic response that cannotbe predicted from individual stress responses. Enrichment analyses of gene ontology terms revealed stress type-specificadjustments of gene expression. Further, global reprogramming mediated by transcription factors and consistent over-representation of basic helix-loop-helix (bHLH) transcription factors, heat shock factors (HSF) and ethylene responsefactors (ERF) was observed. This study reveals the complex transcriptomic responses regulating the perception and signaling ofmultiple abiotic stresses in barley.
You can find more here: https://doi.org/10.1186/s12864-019-5634-0