Insertional mutagenesis is an essential tool to facilitate gene function analysis on a genome-wide scale.
E.g. in maize, the model we focus on, to date only a few hundred of the predicted ~44.000 maize gene models have been functionally characterized. To close this knowledge gap, sequence-indexed reverse genetics collections of loss-of-function mutants for virtually all genes of each genome are required.
While protocols for mutagenesis and flanking sequence tag generation are available for many species and insertion strategies, no tools are accessible for the routine mapping of flanking sequences to genome positions, linking gene annotation to the appropriate seed stocks.
To make loss-of-function mutants for most maize genes more easily available to European researchers, our group recently was part of an effort that introduced a novel Europe based Mutator (Mu) transposon insertional library which was published in Plant Physiology (DOI: https://doi.org/10.1104/pp.20.00478).
As part of this effort and to accelerate the identification, annotation & analysis of Mu insertions we wrote a bioinformatics software designated MuWU, which has since been expanded to be used as a general tool for downstream analysis & annotation of targeted sequencing of transposon insertion sites.
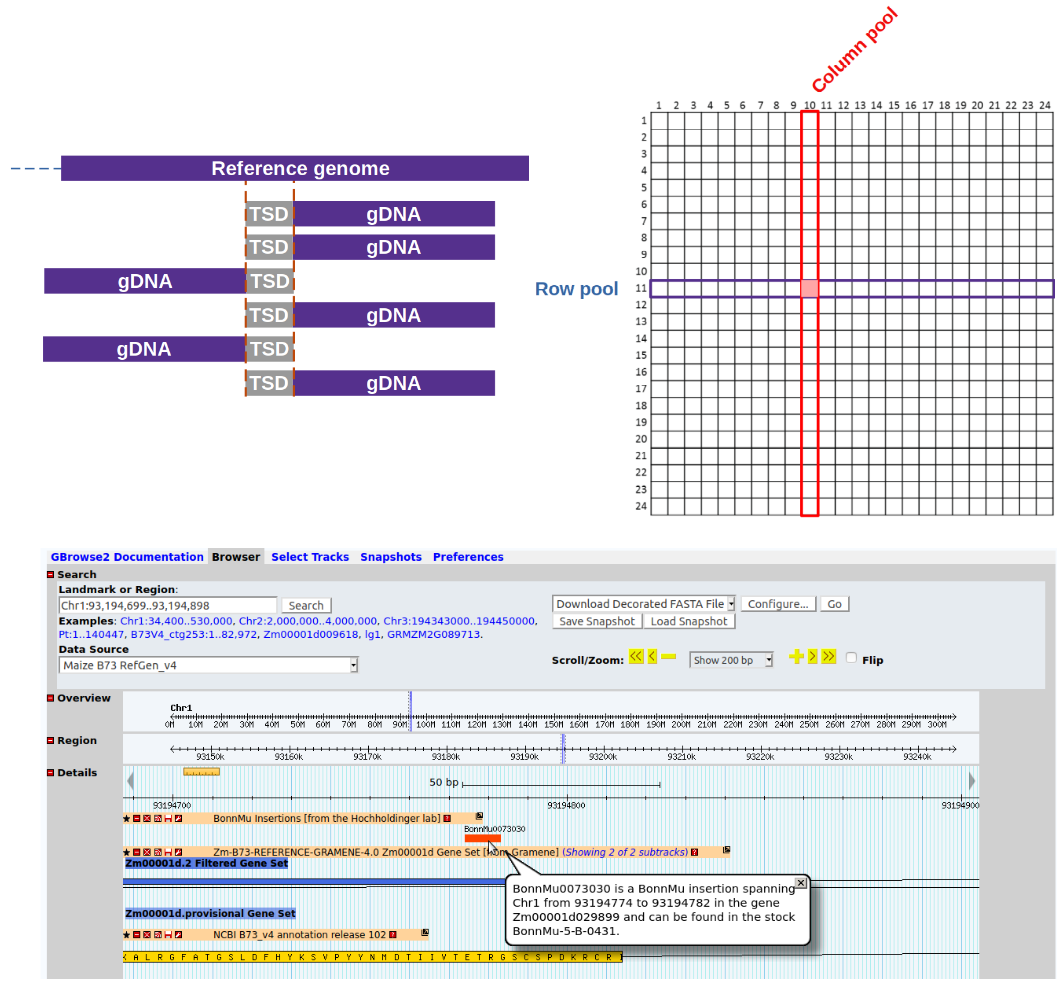
MuWU is a fast pipeline for processing of sequencing reads from an insertional mutagenesis such as Mu-seq. It allows for the reliable identification of tagged genes and is capable of distinguishing between heritable and non-heritable (somatic) insertions/mutations. MuWU further assigns insertions to the corresponding mutated seed stocks which are accessible for researchers around the globe.
Our workflow provides the required robustness and reproducibility needed for routine application, while performing all tasks, from handling of raw sequencing data to creation of final output tables, automatically. It is adaptable to other insertion strategies. To our knowledge, MuWU is the first openly available tool for the analysis of insertional libraries, taking advantage of a multidimensional pooling design and a TE motif of interest to differentiate germinal from somatic insertions.
We believe that our Mu-seq workflow utility will be of broader use to the functional genetics & bioinformatics communities, on the one hand for analysis of insertion libraries, on the other hand as an example of a new generation of custom-designed high throughput sequence data analyses using a workflow approach that is scalable, robust, reproducible and adaptable.
Both the software itself as well as the corresponding publication in Oxford Bioinformatics are published with an open source license and freely available.